Keywords: marginalizing over discretes | mixture model | gaussian mixture model | log-sum-exp trick | pymc3 | Download Notebook
Contents
- The choice of a prior
- The log-sum-exp trick and mixtures
- Pymc3 implements the log-sum-exp directly
- Posterior Predictive
- Marginalizing By Hand
- ADVI
- A much more separated gaussian model
%matplotlib inline
import numpy as np
import scipy as sp
import matplotlib as mpl
import matplotlib.cm as cm
import matplotlib.pyplot as plt
import pandas as pd
pd.set_option('display.width', 500)
pd.set_option('display.max_columns', 100)
pd.set_option('display.notebook_repr_html', True)
import seaborn as sns
sns.set_style("whitegrid")
sns.set_context("poster")
import pymc3 as pm
import theano.tensor as tt
//anaconda/envs/py3l/lib/python3.6/site-packages/h5py/__init__.py:34: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.
from ._conv import register_converters as _register_converters
We’ll first load the tightly coupled gaussian data from the notebook where we did the non-marginalized sampling.
data=np.loadtxt("data/3gv2.dat")
data.shape
(100,)
The choice of a prior
The Dirichlet is the multi-dimensional analog of the Beta. Higher values force you to be more central.
The log-sum-exp trick and mixtures
From the Stan Manual:
\[log\_sum\_exp(a, b) = log(exp(a) + exp(b)).\]The log sum of exponentials function is used to define mixtures on the log scale. It is defined for two inputs by
\[log \left( exp(a) + exp(b) \right) = c + log exp(a − c) + exp(b − c) ,\]If a and b are probabilities on the log scale, then $exp(a) + exp(b)$ is their sum on the linear scale, and the outer log converts the result back to the log scale; to summarize, log_sum_exp does linear addition on the log scale. The reason to use the built-in log_sum_exp function is that it can prevent underflow and overflow in the exponentiation, by calculating the result as
where c = max(a, b). In this evaluation, one of the terms, a − c or b − c, is zero and the other is negative, thus eliminating the possibility of overflow or underflow in the leading term and eking the most arithmetic precision possible out of the operation.
As one can see below, pymc3 uses the same definition
From https://github.com/pymc-devs/pymc3/blob/master/pymc3/math.py#L27
def logsumexp(x, axis=None):
# Adapted from https://github.com/Theano/Theano/issues/1563
x_max = tt.max(x, axis=axis, keepdims=True)
return tt.log(tt.sum(tt.exp(x - x_max), axis=axis, keepdims=True)) + x_max
For example (as taken from the Stan Manual), the mixture of $N(−1, 2)$ and $N(3, 1)$ with mixing proportion $\lambda = (0.3, 0.7)$:
\[logp(y \vert \lambda, \mu, \sigma)\] \[= log\left(0.3×N(y \vert −1,2) + 0.7×N(y \vert 3,1)\right)\] \[= log\left(exp(log(0.3 × N(y \vert − 1, 2))) + exp(log(0.7 × N(y \vert 3, 1))) \right)\] \[= \mathtt{log\_sum\_exp}\left(log(0.3) + log\,N(y \vert − 1, 2), log(0.7) + log\, N(y \vert 3, 1) \right).\]where log_sum_exp is the function as defined above.
This generalizes to the case of more mixture components.
This is thus a custon distribution logp we must define. If we do this, we can go directly from the Dirichlet priors for $p$ and forget the category variable
Pymc3 implements the log-sum-exp directly
Lets see the source here to see how its done:
https://github.com/pymc-devs/pymc3/blob/master/pymc3/distributions/mixture.py
There is a marginalized Gaussian Mixture model available, as well as a general mixture. We’ll use the NormalMixture, to which we must provide mixing weights and components.
import pymc3.distributions.transforms as tr
with pm.Model() as mof3:
p = pm.Dirichlet('p', a=np.array([10., 10., 10.]), shape=3)
means = pm.Normal('means', mu=0, sd=10, shape=3, transform=tr.ordered,
testval=np.array([-1, 0, 1]))
points = pm.NormalMixture('obs', p, mu=means, sd=1, observed=data)
with mof3:
tripletrace_full3 = pm.sample(10000, tune=2000, nuts_kwargs=dict(target_accept=0.95))
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [means, p]
Sampling 2 chains: 100%|██████████| 24000/24000 [01:34<00:00, 253.32draws/s]
The number of effective samples is smaller than 25% for some parameters.
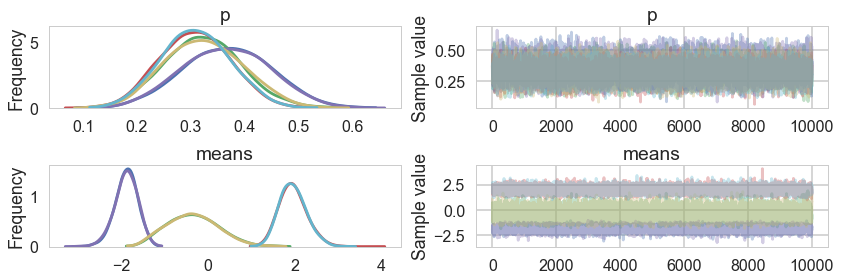
pm.traceplot(tripletrace_full3, combined=True);
//anaconda/envs/py3l/lib/python3.6/site-packages/matplotlib/axes/_base.py:3604: MatplotlibDeprecationWarning:
The `ymin` argument was deprecated in Matplotlib 3.0 and will be removed in 3.2. Use `bottom` instead.
alternative='`bottom`', obj_type='argument')
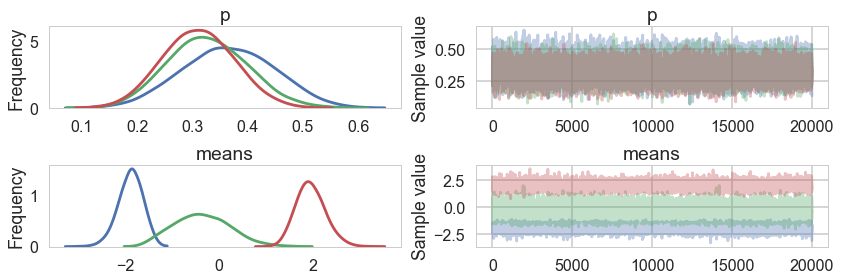
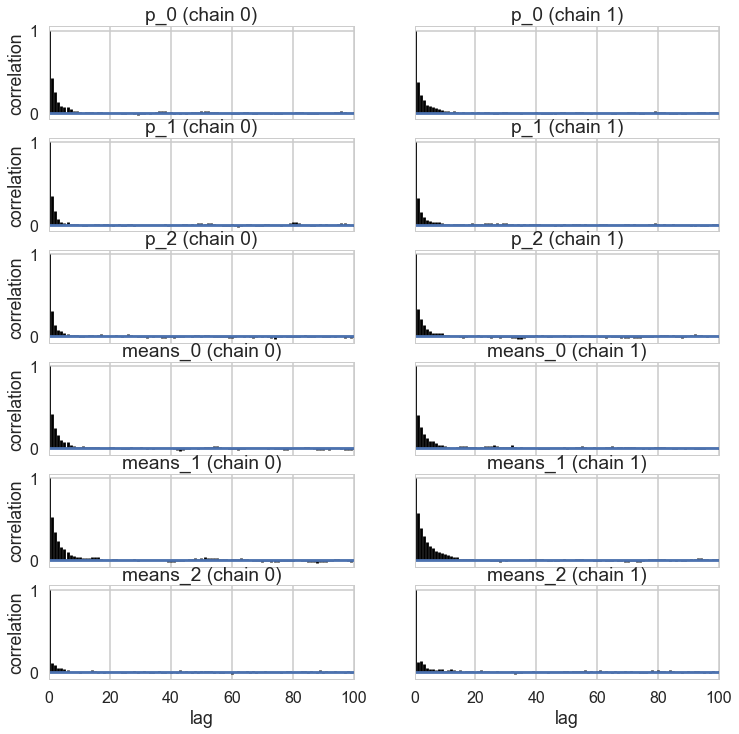
pm.autocorrplot(tripletrace_full3);
pm.plot_posterior(tripletrace_full3);
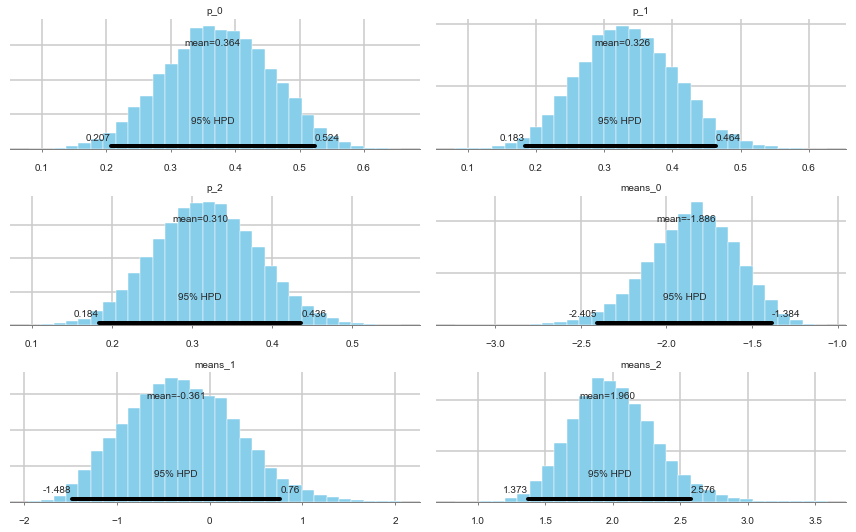
Ordered, even with Dirichlets, our model fits quite nicely.
Posterior Predictive
with mof3:
ppc_trace = pm.sample_ppc(tripletrace_full3, 5000)
100%|██████████| 5000/5000 [00:10<00:00, 479.60it/s]
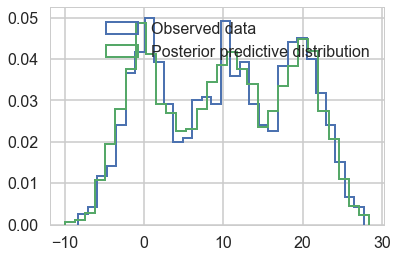
plt.hist(data, bins=30, normed=True,
histtype='step', lw=2,
label='Observed data');
plt.hist(ppc_trace['obs'][:,0], bins=30, normed=True,
histtype='step', lw=2,
label='Posterior predictive distribution');
plt.legend(loc=1);
//anaconda/envs/py3l/lib/python3.6/site-packages/matplotlib/axes/_axes.py:6521: MatplotlibDeprecationWarning:
The 'normed' kwarg was deprecated in Matplotlib 2.1 and will be removed in 3.1. Use 'density' instead.
alternative="'density'", removal="3.1")
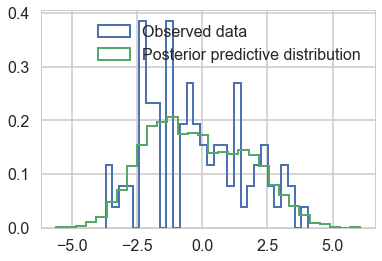
You can see the general agreement between these two distributions in this posterior predictive check!
Marginalizing By Hand
We need to write out the logp for the likelihood ourself now, using logsumexp to do the sum we need.
from pymc3.math import logsumexp
def logp_normal(mu, sigma, value):
# log probability of individual samples
delta = lambda mu: value - mu
return (-1 / 2.) * (tt.log(2 * np.pi) + tt.log(sigma*sigma) +
(delta(mu)* delta(mu))/(sigma*sigma))
# Log likelihood of Gaussian mixture distribution
def logp_gmix(mus, pis, sigmas, n_samples, n_components):
def logp_(value):
logps = [tt.log(pis[i]) + logp_normal(means[i], sigmas[i], value)
for i in range(n_components)]
return tt.sum(logsumexp(tt.stacklists(logps)[:, :n_samples], axis=0))
return logp_
with pm.Model() as mof2:
p = pm.Dirichlet('p', a=np.array([10., 10., 10.]), shape=3)
# cluster centers
means = pm.Normal('means', mu=0, sd=10, shape=3, transform=tr.ordered,
testval=np.array([-1, 0, 1]))
sds = [1., 1., 1.]
# likelihood for each observed valueDensityDist('x', logp_gmix(mus, pi, np.eye(2)), observed=data)
points = pm.DensityDist('obs', logp_gmix(means, p, sds, data.shape[0], 3),
observed=data)
with mof2:
tripletrace_full2 = pm.sample(10000, tune=2000, nuts_kwargs=dict(target_accept=0.95))
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [means, p]
Sampling 2 chains: 100%|██████████| 24000/24000 [01:58<00:00, 202.33draws/s]
The number of effective samples is smaller than 25% for some parameters.
pm.traceplot(tripletrace_full2);
//anaconda/envs/py3l/lib/python3.6/site-packages/matplotlib/axes/_base.py:3604: MatplotlibDeprecationWarning:
The `ymin` argument was deprecated in Matplotlib 3.0 and will be removed in 3.2. Use `bottom` instead.
alternative='`bottom`', obj_type='argument')
Posterior predictive
You cant use sample_ppc directly because we did not create a sampling function for our DensityDist. But this is easy to do for a mixture model. Sample a categorical from the p’s above, and then sample the appropriate gaussian.
Exercise: Write a function to do this!
with mof2:
ppc_trace2 = pm.sample_ppc(tripletrace_full2, 5000)
0%| | 0/5000 [00:00<?, ?it/s]
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
//anaconda/envs/py3l/lib/python3.6/site-packages/pymc3/distributions/distribution.py in _draw_value(param, point, givens, size)
397 try:
--> 398 dist_tmp.random(point=point, size=size)
399 except (ValueError, TypeError):
//anaconda/envs/py3l/lib/python3.6/site-packages/pymc3/distributions/distribution.py in random(self, *args, **kwargs)
212 else:
--> 213 raise ValueError("Distribution was not passed any random method "
214 "Define a custom random method and pass it as kwarg random")
ValueError: Distribution was not passed any random method Define a custom random method and pass it as kwarg random
During handling of the above exception, another exception occurred:
ValueError Traceback (most recent call last)
<ipython-input-45-bc8d116a4478> in <module>()
1 with mof2:
----> 2 ppc_trace2 = pm.sample_ppc(tripletrace_full2, 5000)
//anaconda/envs/py3l/lib/python3.6/site-packages/pymc3/sampling.py in sample_ppc(trace, samples, model, vars, size, random_seed, progressbar)
1127 # draw once to inspect the shape
1128 var_values = list(zip(varnames,
-> 1129 draw_values(vars, point=model.test_point, size=size)))
1130 ppc_trace = defaultdict(list)
1131 for varname, value in var_values:
//anaconda/envs/py3l/lib/python3.6/site-packages/pymc3/distributions/distribution.py in draw_values(params, point, size)
319 else:
320 try: # might evaluate in a bad order,
--> 321 evaluated[param_idx] = _draw_value(param, point=point, givens=givens.values(), size=size)
322 if isinstance(param, collections.Hashable) and named_nodes_parents.get(param):
323 givens[param.name] = (param, evaluated[param_idx])
//anaconda/envs/py3l/lib/python3.6/site-packages/pymc3/distributions/distribution.py in _draw_value(param, point, givens, size)
401 # with theano.shared inputs
402 dist_tmp.shape = np.array([])
--> 403 val = dist_tmp.random(point=point, size=None)
404 dist_tmp.shape = val.shape
405 return dist_tmp.random(point=point, size=size)
//anaconda/envs/py3l/lib/python3.6/site-packages/pymc3/distributions/distribution.py in random(self, *args, **kwargs)
211 return self.rand(*args, **kwargs)
212 else:
--> 213 raise ValueError("Distribution was not passed any random method "
214 "Define a custom random method and pass it as kwarg random")
215
ValueError: Distribution was not passed any random method Define a custom random method and pass it as kwarg random
ADVI
ADVI also needs a marginalized model as it uses gradient descent.
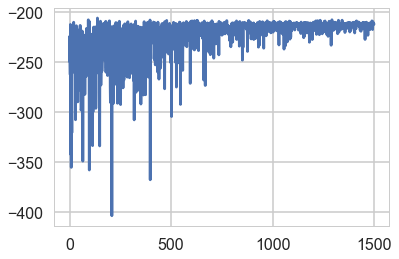
mdvar = pm.ADVI(model=mof3)
mdvar.fit(n=15000)
Average Loss = 213.02: 100%|██████████| 15000/15000 [00:13<00:00, 1081.19it/s]
Finished [100%]: Average Loss = 213.02
<pymc3.variational.approximations.MeanField at 0x1237600b8>
plt.plot(-mdvar.hist[::10])
[<matplotlib.lines.Line2D at 0x125f096a0>]
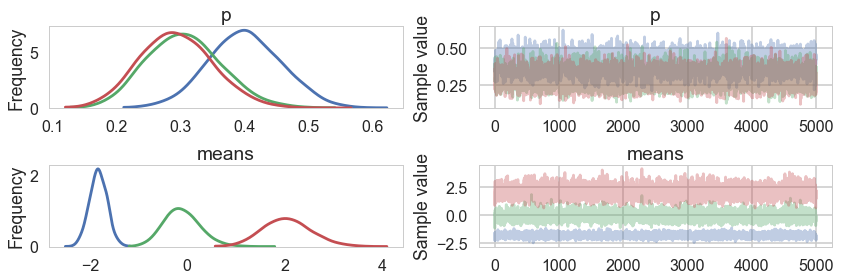
samps=mdvar.approx.sample(5000)
pm.traceplot(samps);
//anaconda/envs/py3l/lib/python3.6/site-packages/matplotlib/axes/_base.py:3604: MatplotlibDeprecationWarning:
The `ymin` argument was deprecated in Matplotlib 3.0 and will be removed in 3.2. Use `bottom` instead.
alternative='`bottom`', obj_type='argument')
pred = pm.sample_ppc(samps, 5000, model=mof3)
100%|██████████| 5000/5000 [00:10<00:00, 463.14it/s]
plt.hist(data, bins=30, normed=True,
histtype='step', lw=2,
label='Observed data');
plt.hist(pred['obs'][:,0], bins=30, normed=True,
histtype='step', lw=2,
label='Posterior predictive distribution');
plt.legend(loc=1);
//anaconda/envs/py3l/lib/python3.6/site-packages/matplotlib/axes/_axes.py:6521: MatplotlibDeprecationWarning:
The 'normed' kwarg was deprecated in Matplotlib 2.1 and will be removed in 3.1. Use 'density' instead.
alternative="'density'", removal="3.1")
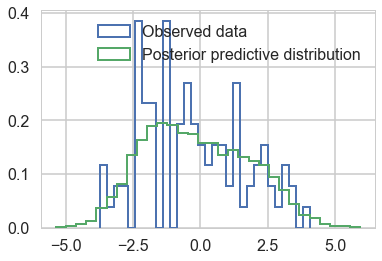
We do quite well!
A much more separated gaussian model
data2=np.loadtxt("data/3g.dat")
plt.hist(data2, bins=30, normed=True,
histtype='step', lw=2,
label='Observed data');
//anaconda/envs/py3l/lib/python3.6/site-packages/matplotlib/axes/_axes.py:6521: MatplotlibDeprecationWarning:
The 'normed' kwarg was deprecated in Matplotlib 2.1 and will be removed in 3.1. Use 'density' instead.
alternative="'density'", removal="3.1")
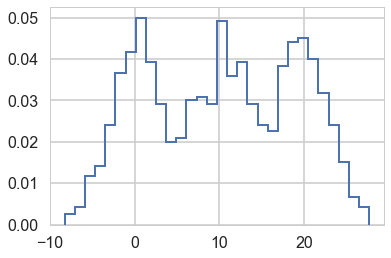
Notice below the use of Potentials. These ensure that there are (a) some members in each cluster and (b) provide us a fifferent way to specify the ordering. This works aby adding log-probability to a model to prevent the sampler going to places where this is negative infinity.
## Class Model for 3 gaussian mixture
with pm.Model() as mofsep:
p = pm.Dirichlet('p', a=np.array([2., 2., 2.]), shape=3)
# ensure all clusters have some points
p_min_potential = pm.Potential('p_min_potential', tt.switch(tt.min(p) < .1, -np.inf, 0))
# cluster centers
means = pm.Normal('means', mu=[0, 10, 20], sd=5, shape=3)
order_means_potential = pm.Potential('order_means_potential',
tt.switch(means[1]-means[0] < 0, -np.inf, 0)
+ tt.switch(means[2]-means[1] < 0, -np.inf, 0))
# measurement error
sds = pm.HalfCauchy('sds', beta=5, shape=3)
# likelihood for each observed value
points = pm.NormalMixture('obs', p, mu=means, sd=sds, observed=data2)
with mofsep:
tracesep_full = pm.sample(10000, tune=2000, nuts_kwargs=dict(target_accept=0.95))
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [sds, means, p]
Sampling 2 chains: 100%|██████████| 24000/24000 [02:09<00:00, 184.97draws/s]
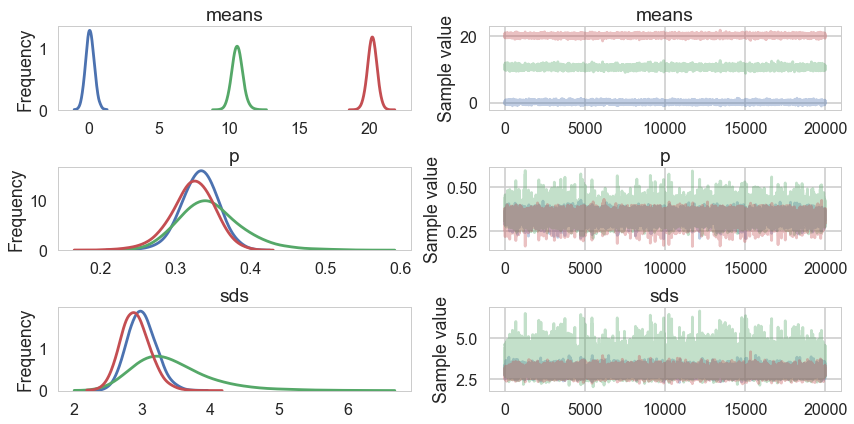
pm.traceplot(tracesep_full, combined=True);
//anaconda/envs/py3l/lib/python3.6/site-packages/matplotlib/axes/_base.py:3604: MatplotlibDeprecationWarning:
The `ymin` argument was deprecated in Matplotlib 3.0 and will be removed in 3.2. Use `bottom` instead.
alternative='`bottom`', obj_type='argument')
with mofsep:
ppc_tracesep = pm.sample_ppc(tracesep_full, 5000)
100%|██████████| 5000/5000 [00:13<00:00, 376.18it/s]
plt.hist(data2, bins=30, normed=True,
histtype='step', lw=2,
label='Observed data');
plt.hist(ppc_tracesep['obs'][:,0], bins=30, normed=True,
histtype='step', lw=2,
label='Posterior predictive distribution');
plt.legend(loc=1);
//anaconda/envs/py3l/lib/python3.6/site-packages/matplotlib/axes/_axes.py:6521: MatplotlibDeprecationWarning:
The 'normed' kwarg was deprecated in Matplotlib 2.1 and will be removed in 3.1. Use 'density' instead.
alternative="'density'", removal="3.1")
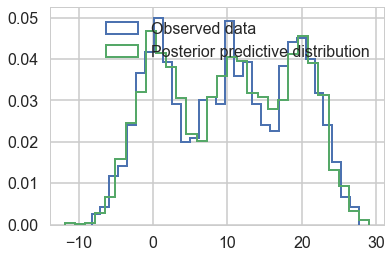
This samples very cleanly and we can do ADVI as well…
mdvarsep = pm.ADVI(model=mofsep)
mdvarsep.fit(n=15000)
Average Loss = 3,475.3: 100%|██████████| 15000/15000 [00:19<00:00, 779.49it/s]
Finished [100%]: Average Loss = 3,475.3
<pymc3.variational.approximations.MeanField at 0x124aca8d0>
plt.plot(-mdvarsep.hist[::10])
[<matplotlib.lines.Line2D at 0x12801ec18>]
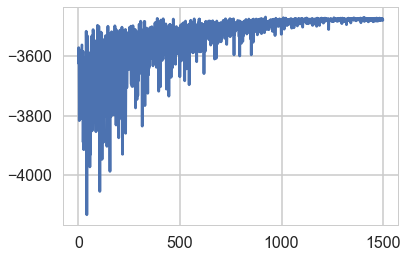
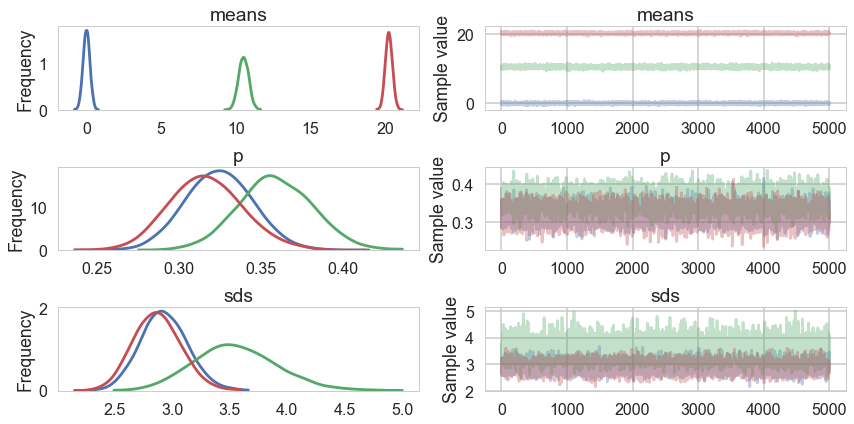
sampssep=mdvarsep.approx.sample(5000)
pm.traceplot(sampssep);
//anaconda/envs/py3l/lib/python3.6/site-packages/matplotlib/axes/_base.py:3604: MatplotlibDeprecationWarning:
The `ymin` argument was deprecated in Matplotlib 3.0 and will be removed in 3.2. Use `bottom` instead.
alternative='`bottom`', obj_type='argument')
predsep = pm.sample_ppc(sampssep, 5000, model=mofsep)
100%|██████████| 5000/5000 [00:11<00:00, 440.11it/s]
plt.hist(data2, bins=30, normed=True,
histtype='step', lw=2,
label='Observed data');
plt.hist(predsep['obs'][:,0], bins=30, normed=True,
histtype='step', lw=2,
label='Posterior predictive distribution');
plt.legend(loc=1);
//anaconda/envs/py3l/lib/python3.6/site-packages/matplotlib/axes/_axes.py:6521: MatplotlibDeprecationWarning:
The 'normed' kwarg was deprecated in Matplotlib 2.1 and will be removed in 3.1. Use 'density' instead.
alternative="'density'", removal="3.1")